Note
This page was generated from mimic_2_causal_inference.ipynb. Some tutorial content may look better in light mode.
MIMIC-II Causal Inference#
This tutorial continues the exploration the MIMIC-II IAC dataset using causal inference methods. The dataset was created for the purpose of a case study in the book: Secondary Analysis of Electronic Health Records, published by Springer in 2016. In particular, the dataset was used throughout Chapter 16 (Data Analysis) by Raffa J. et al. to investigate the effectiveness of indwelling arterial catheters in hemodynamically stable patients with respiratory failure for mortality outcomes. The dataset is derived from MIMIC-II, the publicly-accessible critical care database. It contains summary clinical data and outcomes for 1,776 patients.
Reference:
[1] https://github.com/py-why/dowhy
[2] https://www.pywhy.org/dowhy/
[3] https://arxiv.org/abs/2011.04216
[4] https://physionet.org/content/mimic2-iaccd/1.0/
Importing ehrapy and setting plotting parameters#
[1]:
import ehrapy as ep
from IPython.display import display
import graphviz
[2]:
import warnings
warnings.filterwarnings("ignore")
[3]:
ep.print_versions()
----- ehrapy 0.5.0 rich NA session_info 1.0.0 ----- Levenshtein 0.20.9 PIL 9.4.0 anndata 0.9.1 appnope 0.1.3 astor 0.8.1 asttokens NA attr 23.1.0 autograd NA autograd_gamma NA backcall 0.2.0 beta_ufunc NA binom_ufunc NA bottleneck 1.3.5 bs4 4.12.2 cachetools 5.3.1 category_encoders 2.6.0 certifi 2023.05.07 cffi 1.15.1 charset_normalizer 2.1.1 cloudpickle 2.2.1 cycler 0.10.0 cython_runtime NA dateutil 2.8.2 db_dtypes 1.1.1 debugpy 1.6.7 decorator 5.1.1 deep_translator 1.9.1 deepl 1.14.0 defusedxml 0.7.1 dill 0.3.6 dot_parser NA dowhy 0.9.1 econml 0.14.0 entrypoints 0.4 executing 1.2.0 fhiry 3.0.0 formulaic 0.6.1 future 0.18.3 google NA graphlib NA graphviz 0.20.1 grpc 1.54.2 grpc_status NA h5py 3.8.0 hypergeom_ufunc NA idna 3.4 igraph 0.10.4 interface_meta 1.3.0 ipykernel 6.15.0 ipython_genutils 0.2.0 ipywidgets 8.0.7 jedi 0.18.2 joblib 1.2.0 kiwisolver 1.4.4 leidenalg 0.9.1 lifelines 0.27.7 llvmlite 0.39.1 lxml 4.9.3 matplotlib 3.7.1 missingno 0.5.2 mpl_toolkits NA mpmath 1.3.0 natsort 8.3.1 nbinom_ufunc NA ncf_ufunc NA networkx 2.8.8 numba 0.56.4 numexpr 2.8.4 numpy 1.23.5 packaging 23.0 pandas 1.5.3 parso 0.8.3 patsy 0.5.3 pexpect 4.8.0 pickleshare 0.7.5 pkg_resources NA prompt_toolkit 3.0.38 psutil 5.9.5 ptyprocess 0.7.0 pure_eval 0.2.2 pyarrow 12.0.0 pyasn1 0.5.0 pyasn1_modules 0.3.0 pydev_ipython NA pydevconsole NA pydevd 2.9.5 pydevd_file_utils NA pydevd_plugins NA pydevd_tracing NA pydot 1.4.1 pygments 2.15.1 pyparsing 3.0.9 pytz 2023.3 rapidfuzz 2.15.1 requests 2.31.0 rsa 4.9 scanpy 1.9.3 scipy 1.9.3 seaborn 0.12.2 setuptools 67.8.0 six 1.16.0 sklearn 1.1.3 soupsieve 2.4.1 sparse 0.14.0 sphinxcontrib NA stack_data 0.6.2 statsmodels 0.13.5 sympy 1.11.1 texttable 1.6.7 thefuzz 0.19.0 threadpoolctl 3.1.0 torch 2.0.1 tornado 6.2 tqdm 4.65.0 traitlets 5.9.0 typing_extensions NA urllib3 2.0.3 wcwidth 0.2.6 wrapt 1.15.0 yaml 6.0 zmq 25.0.2 zoneinfo NA ----- IPython 8.8.0 jupyter_client 7.4.9 jupyter_core 4.11.2 notebook 6.4.12 ----- Python 3.10.11 (main, Apr 20 2023, 13:58:42) [Clang 14.0.6 ] macOS-13.1-arm64-arm-64bit ----- Session information updated at 2023-07-27 13:27
MIMIC-II dataset preparation#
Let’s load the MIMIC-II dataset using ehrapy with default one-hot encoding.
[4]:
adata = ep.dt.mimic_2(encoded=True)
2023-07-27 13:27:20,502 - root INFO - Added all columns to `obs`.
2023-07-27 13:27:20,513 - root INFO - Transformed passed dataframe into an AnnData object with n_obs x n_vars = `1776` x `46`.
2023-07-27 13:27:20,514 - root INFO - The original categorical values `['day_icu_intime', 'service_unit']` were added to uns.
2023-07-27 13:27:20,524 - root INFO - Encoding strings in X to save to .h5ad. Loading the file will reverse the encoding.
2023-07-27 13:27:20,526 - root INFO - Updated the original layer after encoding.
2023-07-27 13:27:20,535 - root INFO - The original categorical values `['day_icu_intime', 'service_unit']` were added to obs.
The MIMIC-II dataset has 1776 patients as described above with 46 features.
[5]:
adata
[5]:
AnnData object with n_obs × n_vars = 1776 × 46
obs: 'service_unit', 'day_icu_intime'
uns: 'numerical_columns', 'non_numerical_columns', 'original_values_categoricals', 'var_to_encoding', 'encoding_to_var'
layers: 'original'
Causal Inference on the MIMIC-II dataset#
In the background, ehrapy uses the dowhy package to enable effortless causal inference on electronic health records (EHR). Any dowhy analysis is structured into 3 steps: 1) Formulate causal questions 2) Estimate causal effects 3) Perform refutation tests.
Within ehrapy, we have consolidated this method into a single function ehrapy.causal_inference(). This function takes in the dataset, the treatment variable, the outcome variable, and the further optional parameters. It then performs the 3 steps above and displays the information in a user-friendly way.
Causal Graph#
The causal graph is a directed acyclic graph (DAG) that represents the causal relationships between the variables in the dataset. Here, we create it by manually writing out the connections between the variables. Other options would be the GML or DOT graph format. Furthermore, you can use graphical tools like DAGitty to construct the graph. You can export the graph string that it generates. The graph string is very close to the DOT format: just rename dag to digraph, remove newlines and add a semicolon after every line, to convert it to the DOT format and input to DoWhy.
Assumptions:#
both age and overweight increase your risk for medical problems
having a lot of problems makes you more likely to die in the hospital
having a lot of problems influences your likelihood of getting an IAC
having an IAC influences your likelihood of dying in the hospital
[6]:
causal_graph = """digraph {
aline_flg[label="Indwelling arterial catheters used"];
icu_los_day[label="Days in ICU"];
age -> sepsis_flg;
age -> chf_flg;
age -> afib_flg;
age -> renal_flg;
age -> liver_flg;
age -> copd_flg;
age -> cad_flg;
age -> stroke_flg;
age -> resp_flg;
bmi -> sepsis_flg;
bmi -> chf_flg;
bmi -> afib_flg;
bmi -> renal_flg;
bmi -> liver_flg;
bmi -> copd_flg;
bmi -> cad_flg;
bmi -> stroke_flg;
bmi -> resp_flg;
sepsis_flg -> aline_flg;
chf_flg -> aline_flg;
afib_flg -> aline_flg;
renal_flg -> aline_flg;
liver_flg -> aline_flg;
copd_flg -> aline_flg;
cad_flg -> aline_flg;
stroke_flg -> aline_flg;
resp_flg -> aline_flg;
sepsis_flg -> icu_los_day;
chf_flg -> icu_los_day;
afib_flg -> icu_los_day;
renal_flg -> icu_los_day;
liver_flg -> icu_los_day;
copd_flg -> icu_los_day;
cad_flg -> icu_los_day;
stroke_flg -> icu_los_day;
resp_flg -> icu_los_day;
aline_flg -> icu_los_day;
}"""
g = graphviz.Source(causal_graph)
display(g)
Causal inference#
This graph can now be fed into the causal_inference() method. Furthermore, we have to specify an estimation_method. For now, we will use backdoor.linear_regression. Please refer to this example notebook and the official dowhy documentation for more information on the different estimation methods.
[7]:
_ = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
)
2023-07-27 13:27:22,813 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (1/10)
2023-07-27 13:27:24,597 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (2/10)
2023-07-27 13:27:26,725 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (3/10)
2023-07-27 13:27:28,374 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (4/10)
2023-07-27 13:27:30,247 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (5/10)
2023-07-27 13:27:31,896 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (6/10)
2023-07-27 13:27:33,888 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (7/10)
2023-07-27 13:27:36,207 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (8/10)
2023-07-27 13:27:37,698 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (9/10)
2023-07-27 13:27:39,433 - root WARNING - [dowhy] Refutation 'placebo_treatment_refuter' returned invalid pval 'nan', retrying (10/10)
Causal inference results for treatment variable 'aline_flg' and outcome variable 'icu_los_day':
└- Increasing the treatment variable(s) [aline_flg] from 0 to 1 causes an increase of 2.234881309168317 in the expected value of the outcome [icu_los_day], over the data distribution/population represented by the dataset.
Refutation results
├-Refute: Use a Placebo Treatment
| ├- Estimated effect: 2.23
| ├- New effect: 0.000
| ├- p-value: nan
| └- Test significance: 2.23
├-Refute: Add a random common cause
| ├- Estimated effect: 2.23
| ├- New effect: 2.234
| ├- p-value: 0.435
| └- Test significance: 2.23
├-Refute: Use a subset of data
| ├- Estimated effect: 2.23
| ├- New effect: 2.244
| ├- p-value: 0.445
| └- Test significance: 2.23
└-Refute: Add an Unobserved Common Cause
├- Estimated effect: 2.23
├- New effect: -0.52, 2.33
├- p-value: Not applicable
└- Test significance: 2.23
As we can see, the model returns a summary of the identified causal effect and the refutation results. The placebo_treatment_refuter failed in this case because our intervention variable is binary. Per default, all 4 potential refutation methods are run, but the user can also specify to only use a subset of them.
[8]:
_ = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
)
Causal inference results for treatment variable 'aline_flg' and outcome variable 'icu_los_day':
└- Increasing the treatment variable(s) [aline_flg] from 0 to 1 causes an increase of 2.234881309168317 in the expected value of the outcome [icu_los_day], over the data distribution/population represented by the dataset.
Refutation results
├-Refute: Add a random common cause
| ├- Estimated effect: 2.23
| ├- New effect: 2.235
| ├- p-value: 0.494
| └- Test significance: 2.23
├-Refute: Use a subset of data
| ├- Estimated effect: 2.23
| ├- New effect: 2.231
| ├- p-value: 0.478
| └- Test significance: 2.23
└-Refute: Add an Unobserved Common Cause
├- Estimated effect: 2.23
├- New effect: -0.44, 2.29
├- p-value: Not applicable
└- Test significance: 2.23
By default, we are hiding a lot of the default dowhy output for clarity. However, it is possible to nontheless display it.
[9]:
_ = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
print_causal_estimate=True,
print_summary=False,
)
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
────────────(E[icu_los_day|resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,re
d[aline_flg]
nal_flg,liver_flg,chf_flg,cad_flg])
Estimand assumption 1, Unconfoundedness: If U→{aline_flg} and U→icu_los_day then P(icu_los_day|aline_flg,resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,renal_flg,liver_flg,chf_flg,cad_flg,U) = P(icu_los_day|aline_flg,resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,renal_flg,liver_flg,chf_flg,cad_flg)
## Realized estimand
b: icu_los_day~aline_flg+resp_flg+stroke_flg+afib_flg+copd_flg+sepsis_flg+renal_flg+liver_flg+chf_flg+cad_flg
Target units: ate
## Estimate
Mean value: 2.234881309168317
Plotting#
Furthermore, the causal_inference() function can generate several plots, if desired. It’s for example possible to output the causal graph …
[10]:
_ = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
print_causal_estimate=False,
print_summary=False,
show_graph=True,
)
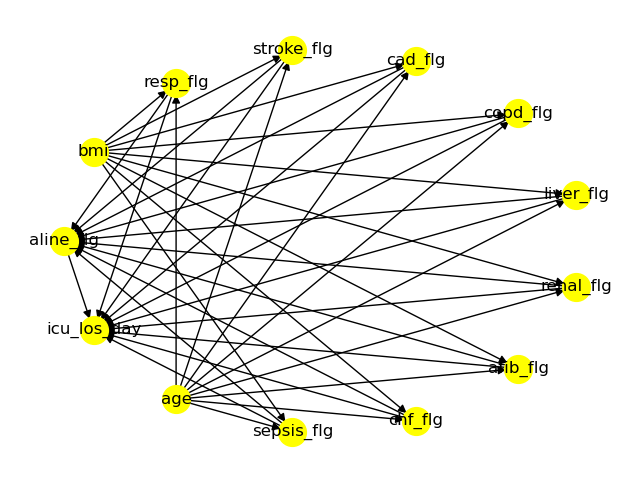
… or the graph generated by the add_unobserved_common_cause refutation method.
[11]:
_ = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
print_causal_estimate=False,
print_summary=False,
show_graph=False,
show_refute_plots=True,
)
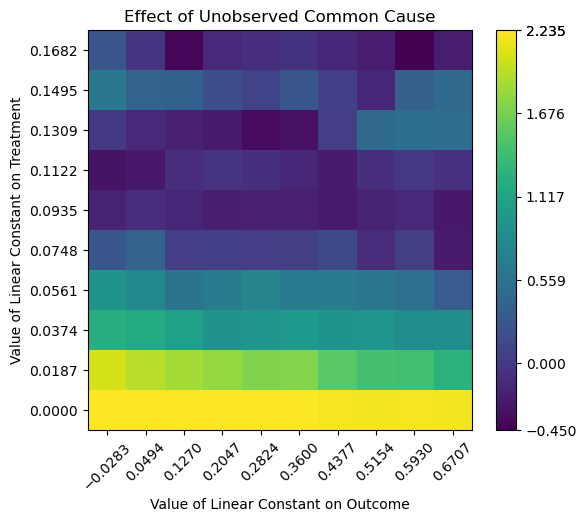
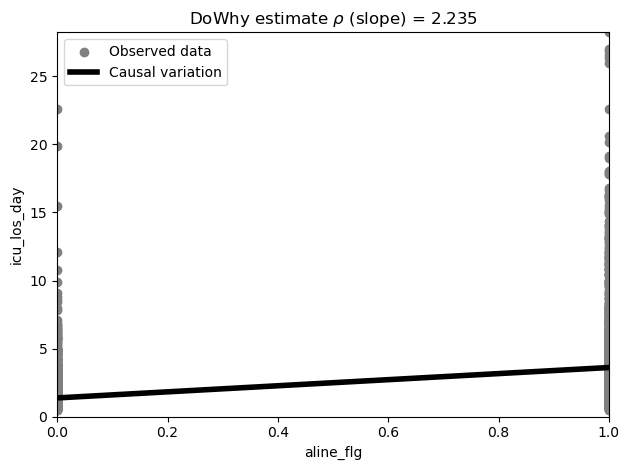
In the process of performing the causal dowhy analysis, an estimator is created which we can query from the model and then visualise.
[12]:
estimate = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
print_causal_estimate=False,
print_summary=False,
show_graph=False,
show_refute_plots=False,
return_as="estimate",
)
[13]:
ep.pl.causal_effect(estimate)
[13]:
<Axes: title={'center': 'DoWhy estimate $\\rho$ (slope) = 2.235'}, xlabel='aline_flg', ylabel='icu_los_day'>
Advanced options#
Within dowhy, the user can specify several arguments for identification, estimation and refutation. These arguments can also be passed directly to the respective functions through the identify_kwargs, estimate_kwargs and refute_kwarg of causal_inference().
[14]:
estimate = ep.tl.causal_inference(
adata=adata,
graph=causal_graph,
treatment="aline_flg",
outcome="icu_los_day",
estimation_method="backdoor.linear_regression",
refute_methods=[
# "placebo_treatment_refuter", # we know it'll fail
"random_common_cause",
"data_subset_refuter",
"add_unobserved_common_cause",
],
print_causal_estimate=True,
print_summary=True,
show_graph=False,
show_refute_plots="contour",
return_as="estimate",
identify_kwargs={"proceed_when_unidentifiable": True},
estimate_kwargs={"target_units": "days"},
refute_kwargs={"random_seed": 5},
)
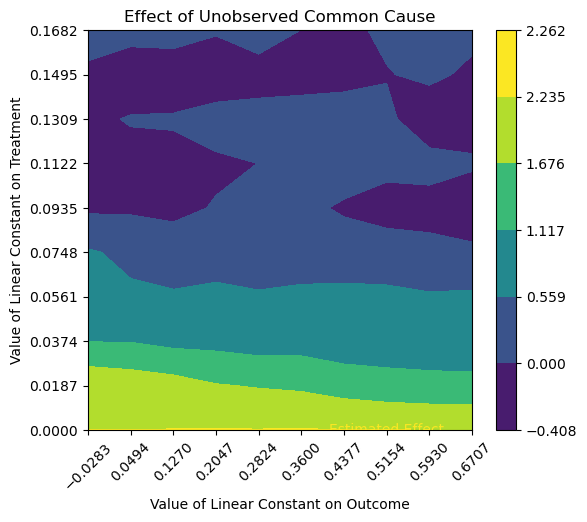
*** Causal Estimate ***
## Identified estimand
Estimand type: EstimandType.NONPARAMETRIC_ATE
### Estimand : 1
Estimand name: backdoor
Estimand expression:
d
────────────(E[icu_los_day|resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,re
d[aline_flg]
nal_flg,liver_flg,chf_flg,cad_flg])
Estimand assumption 1, Unconfoundedness: If U→{aline_flg} and U→icu_los_day then P(icu_los_day|aline_flg,resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,renal_flg,liver_flg,chf_flg,cad_flg,U) = P(icu_los_day|aline_flg,resp_flg,stroke_flg,afib_flg,copd_flg,sepsis_flg,renal_flg,liver_flg,chf_flg,cad_flg)
## Realized estimand
b: icu_los_day~aline_flg+resp_flg+stroke_flg+afib_flg+copd_flg+sepsis_flg+renal_flg+liver_flg+chf_flg+cad_flg
Target units: days
## Estimate
Mean value: 2.234881309168317
Causal inference results for treatment variable 'aline_flg' and outcome variable 'icu_los_day':
└- Increasing the treatment variable(s) [aline_flg] from 0 to 1 causes an increase of 2.234881309168317 in the expected value of the outcome [icu_los_day], over the data distribution/population represented by the dataset.
Refutation results
├-Refute: Add a random common cause
| ├- Estimated effect: 2.23
| ├- New effect: 2.235
| ├- p-value: 0.484
| └- Test significance: 2.23
├-Refute: Use a subset of data
| ├- Estimated effect: 2.23
| ├- New effect: 2.234
| ├- p-value: 0.495
| └- Test significance: 2.23
└-Refute: Add an Unobserved Common Cause
├- Estimated effect: 2.23
├- New effect: -0.41, 2.26
├- p-value: Not applicable
└- Test significance: 2.23