Note
This page was generated from medcat.ipynb. Some tutorial content may look better in light mode.
Extracting from free text with MedCAT#
This tutorial serves as an introduction on how to use ehrapy together with MedCAT. MedCat is a tool to extract medical entities from free text and link it to biomedical ontologies. Biomedical entities could be anything biomedical; not only diagnoses or diseases but also symptoms, drugs or even peptides. It also tries to keep the context of an extracted entitiy (for example, whether a specific disease has been diagnosed or not). This is especially important for electronic health records data, as most of the time doctors notes are simply copied and pasted into the data and not preprocessed in any form. Consider the following example:
The patient suffers from diabetes.
vs.
The patient does not suffer from diabetes.
In detail, ehrapy uses a pretrained and packages model from MedCat (https://medcat.readthedocs.io/en/latest/main.html#models). This model is limited in performance but good enough for this demonstration. A larger (trained) model is planned to be released somewhen in the (near) future by the MedCAT maintainers.
[1]:
import ehrapy as ep
import pandas as pd
[2]:
ep.settings.n_jobs = 2
Download the example data
[3]:
!wget -nc https://raw.githubusercontent.com/CogStack/MedCATtutorials/main/notebooks/introductory/data/pt_notes.csv -P ./medcat_data/
!wget -nc https://medcat.rosalind.kcl.ac.uk/media/medmen_wstatus_2021_oct.zip -P ./medcat_data/
File ‘./medcat_data/pt_notes.csv’ already there; not retrieving.
File ‘./medcat_data/medmen_wstatus_2021_oct.zip’ already there; not retrieving.
Custom MedCAT object of ehrapy#
To allow for seemless interoperability of ehrapy and MedCAT, we require a “superobject” that references the AnnData object while providing the MedCAT functionality. This object further stores MedCAT related features such as a vocabulary, a concept database and (later on in this tutorial) also the annotated results.
First, we read the example data into an AnnData object, encode the data and calculate neighbors for subsequent processing. As a next step, we create the MedCAT object with a modelpack and set default TUI filters. A full list of TUI’s can be found at: https://lhncbc.nlm.nih.gov/ii/tools/MetaMap/Docs/SemanticTypes_2018AB.txt
[4]:
adata = ep.io.read_csv("medcat_data/pt_notes.csv", columns_obs_only=["text"])
adata_encoded = ep.pp.encode(adata, autodetect=True)
ep.pp.neighbors(adata_encoded)
# create the main ehrapy medcat object used for medical entitiy analysis using ehrapy and medcat
ep_medcat = ep.tl.MedCAT(
adata_encoded, model_pack_path="./medcat_data/medmen_wstatus_2021_oct.zip"
)
# only use diseases and behavioural disorders diagnoses in this example by filtering by TUI
ep_medcat.set_filter_by_tui(tuis=["T047", "T048"])
2023-08-10 17:54:04,813 - root INFO - Transformed passed dataframe into an AnnData object with n_obs x n_vars = `1088` x `8`.
2023-08-10 17:54:04,814 - root INFO - The original categorical values `['gender', 'category']` were added to uns.
2023-08-10 17:54:04,832 - root INFO - Encoding strings in X to save to .h5ad. Loading the file will reverse the encoding.
2023-08-10 17:54:04,837 - root INFO - Updated the original layer after encoding.
2023-08-10 17:54:04,853 - root INFO - The original categorical values `['gender', 'category']` were added to obs.
Using this model pack we can already extract entities from an example note.
[5]:
text = "He was diagnosed with kidney failure"
doc = ep_medcat.cat(text)
doc.ents
[5]:
(kidney failure,)
[6]:
# Example output of an extracted medcat entity; note that ehrapy will deal with this automatically and the here displayed manual extraction is not required.
# CUI: Concept Unique Identifier, which is just an unique identifier for each concept extracted
ep_medcat.cat.get_entities("He was diagnosed with kidney failure", only_cui=False)
[6]:
{'entities': {2: {'pretty_name': 'Kidney Failure',
'cui': 'C0035078',
'type_ids': ['T047'],
'types': ['Disease or Syndrome'],
'source_value': 'kidney failure',
'detected_name': 'kidney~failure',
'acc': 1.0,
'context_similarity': 1.0,
'start': 22,
'end': 36,
'icd10': [],
'ontologies': [],
'snomed': [],
'id': 2,
'meta_anns': {'Status': {'value': 'Affirmed',
'confidence': 0.9999961853027344,
'name': 'Status'}}}},
'tokens': []}
Extracting and visualizing all disease entities#
To extract all disease entities from our example dataset we require a complete annotation of the dataset. This step is computationally expensive and may take some time.
[7]:
ep.tl.mc.annotate_text(ep_medcat, text_column="text", n_proc=2)
The annotated results as extracted by MedCAT are transformed and stored into a Pandas DataFrame
[8]:
ep_medcat.annotated_results
[8]:
| row_nr | pretty_name | cui | type_ids | types | meta_anns | |
|---|---|---|---|---|---|---|
| 0 | 0 | Degenerative polyarthritis | C0029408 | [T047] | [Disease or Syndrome] | Affirmed |
| 1 | 0 | Atrial Fibrillation | C0004238 | [T047] | [Disease or Syndrome] | Affirmed |
| 2 | 0 | Hypertensive disease | C0020538 | [T047] | [Disease or Syndrome] | Affirmed |
| 3 | 0 | Asthma | C0004096 | [T047] | [Disease or Syndrome] | Affirmed |
| 4 | 0 | Sleep Apnea, Obstructive | C0520679 | [T047] | [Disease or Syndrome] | Affirmed |
| ... | ... | ... | ... | ... | ... | ... |
| 10781 | 999 | Diabetes | C0011847 | [T047] | [Disease or Syndrome] | Other |
| 10782 | 999 | Diabetes | C0011847 | [T047] | [Disease or Syndrome] | Affirmed |
| 10783 | 999 | Diabetes | C0011847 | [T047] | [Disease or Syndrome] | Affirmed |
| 10784 | 999 | Diabetes Mellitus | C0011849 | [T047] | [Disease or Syndrome] | Affirmed |
| 10785 | 999 | Diabetes | C0011847 | [T047] | [Disease or Syndrome] | Affirmed |
10786 rows × 6 columns
We can also get a proper overview for the top 10 most entities found in the data (affirmed diagnoses only).
[9]:
ep.tl.mc.get_annotation_overview(ep_medcat)
pretty_name type_ids types n_patient_visit n_patient_visit_percent ───────────────────────────────────────────────────────────────────────────────────────────────────────────────── Hypertensive disease T047 Disease or Syndrome 432 42.39450441609421 Obesity T047 Disease or Syndrome 245 24.043179587831208 Diabetes T047 Disease or Syndrome 149 14.622178606476938 Coronary Arteriosclerosis T047 Disease or Syndrome 127 12.463199214916584 Diabetes Mellitus T047 Disease or Syndrome 118 11.579980372914623 Disease T047 Disease or Syndrome 110 10.794896957801766 Cerebrovascular accident T047 Disease or Syndrome 110 10.794896957801766 Asthma T047 Disease or Syndrome 97 9.519136408243375 Chronic Obstructive Airway T047 Disease or Syndrome 92 9.028459273797841 Disease Mental Depression T048 Mental or Behavioral 91 8.930323846908735 Dysfunction
Alternatively, we can also get an overview of the top 10 negated diagnoses only.
[10]:
ep.tl.mc.get_annotation_overview(ep_medcat, status="Other")
pretty_name type_ids types n_patient_visit n_patient_visit_percent ───────────────────────────────────────────────────────────────────────────────────────────────────────────────── Cerebrovascular accident T047 Disease or Syndrome 119 15.973154362416109 Erythema T047 Disease or Syndrome 85 11.409395973154362 Diabetes T047 Disease or Syndrome 78 10.46979865771812 Hypertensive disease T047 Disease or Syndrome 68 9.12751677852349 disorder lesions skin T047 Disease or Syndrome 68 9.12751677852349 Lymphadenopathy T047 Disease or Syndrome 64 8.590604026845638 nervous system disorder T047 Disease or Syndrome 58 7.785234899328859 Coronary Arteriosclerosis T047 Disease or Syndrome 50 6.7114093959731544 Drug abuse T048 Mental or Behavioral 50 6.7114093959731544 Dysfunction Pneumonia T047 Disease or Syndrome 50 6.7114093959731544
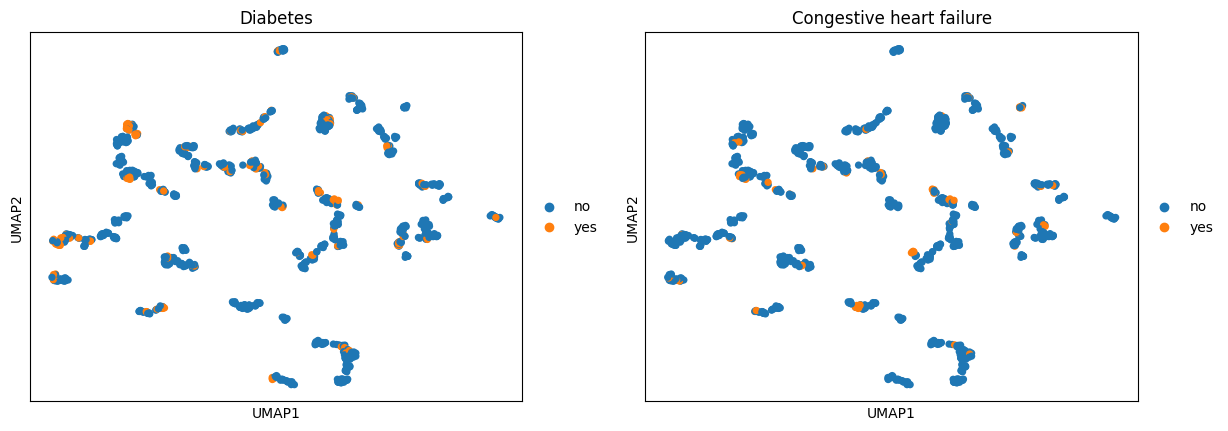
Since our MedCAT is aware of both, the annotation results and the AnnData object, we can just pass it to any plotting functions of ehrapy, just we like we are used to with AnnData objects. First, we calculate a UMAP embedding using the AnnData object and then we color by all patients that had some form of Pneumonia.
[11]:
ep.tl.umap(ep_medcat.anndata)
ep.pl.umap(ep_medcat, color=["Diabetes", "Congestive heart failure"])
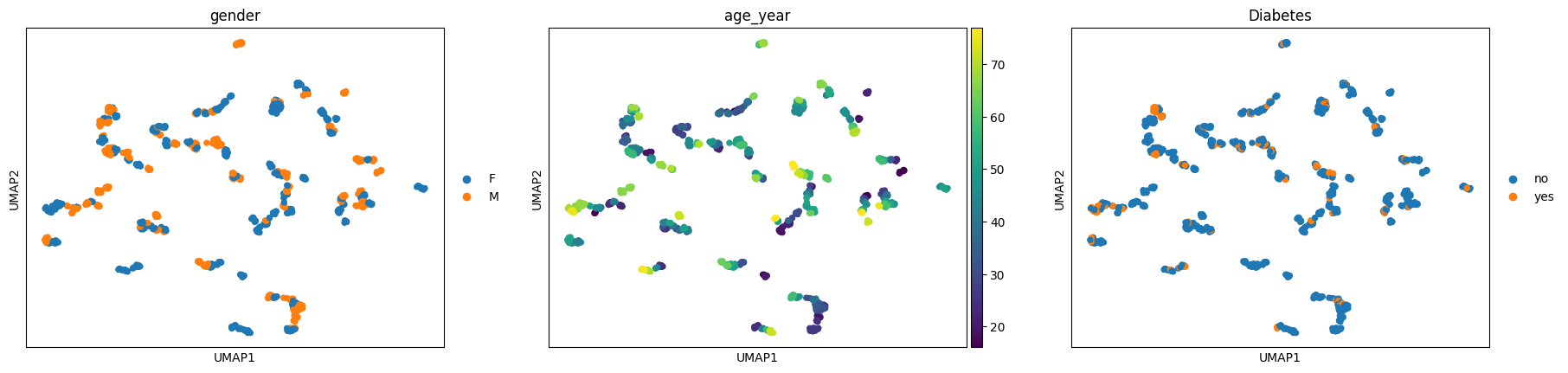
It is still possible to color by the original columns in the AnnData object with the MedCAT object.
[12]:
ep.pl.umap(ep_medcat, color=["gender", "age_year", "Diabetes"])
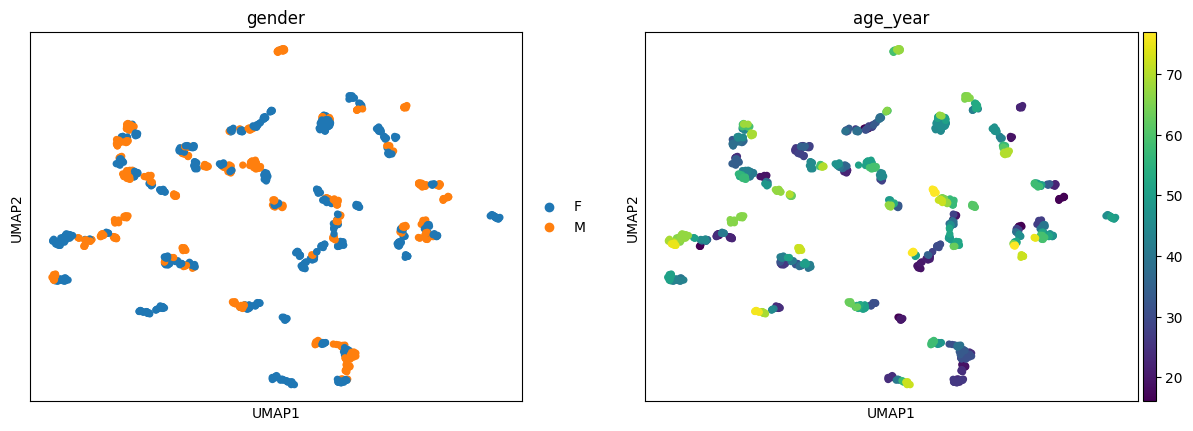
Passing an AnnData object instead of an ehrapy MedCAT object will also work, but not with extracted entities.
[13]:
ep.pl.umap(ep_medcat.anndata, color=["gender", "age_year"])
Typos are automatically fixed by ehrapy whenever possible.
[14]:
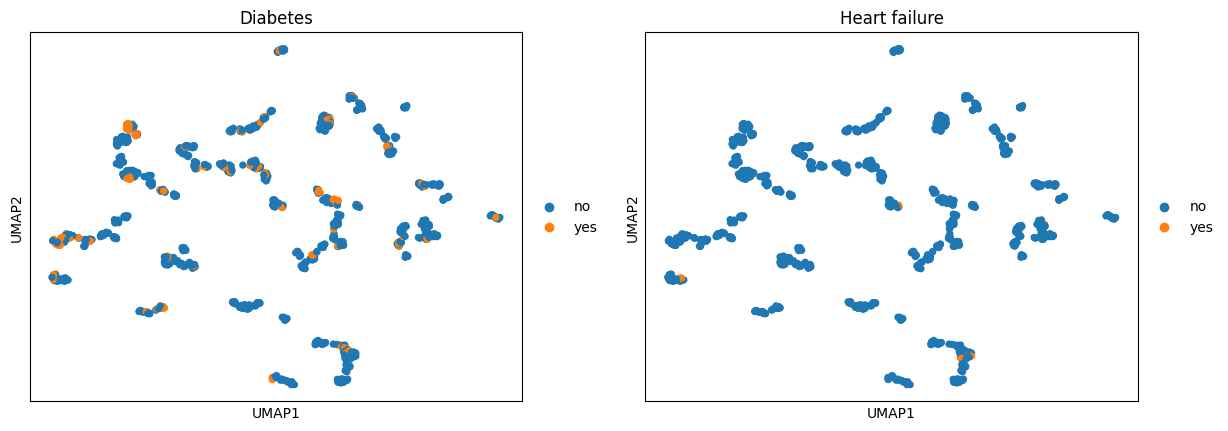
ep.pl.umap(ep_medcat, color=["Diubetes", "Heart Failure"])
Did not find Diubetes in MedCAT's extracted entities. Will use best match Diabetes!
Did not find Heart Failure in MedCAT's extracted entities. Will use best match Heart failure!
All other features of ehrapy are of course also available.
[15]:
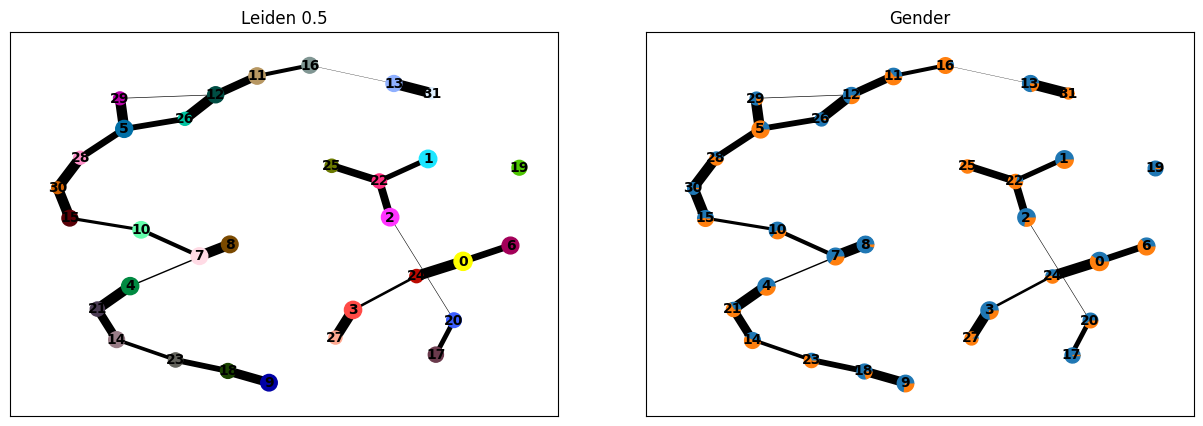
ep.tl.leiden(ep_medcat.anndata, resolution=0.5, key_added="leiden_0_5")
ep.tl.paga(ep_medcat.anndata, groups="leiden_0_5")
# paga currently does not support medcat extracted entities directly (as of ehrapy 0.2.0)
ep.pl.paga(
ep_medcat.anndata,
color=["leiden_0_5", "gender"],
cmap=ep.pl.Colormaps.grey_red.value,
title=["Leiden 0.5", "Gender"],
)
[16]:
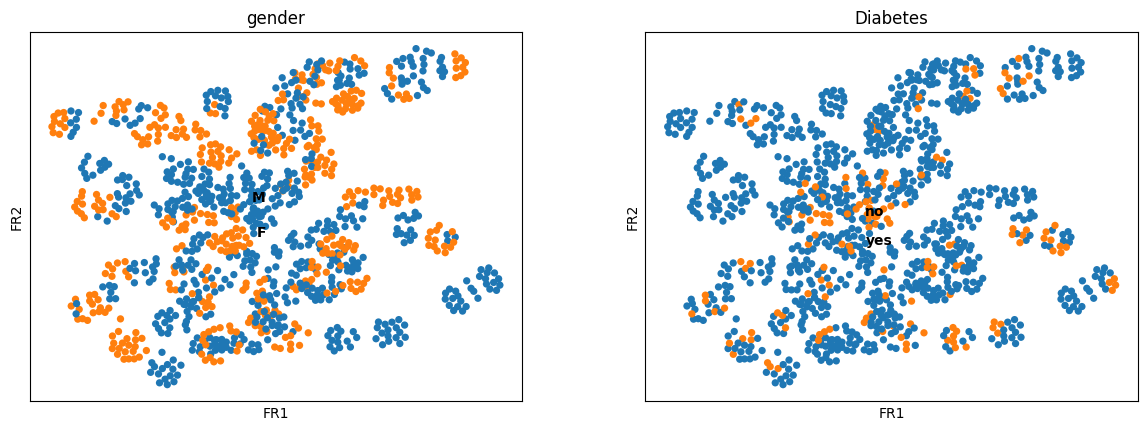
ep.tl.draw_graph(ep_medcat.anndata, init_pos="paga")
ep.pl.draw_graph(ep_medcat, color=["gender", "Diabetes"], legend_loc="on data")
WARNING: Package 'fa2' is not installed, falling back to layout 'fr'.To use the faster and better ForceAtlas2 layout, install package 'fa2' (`pip install fa2`).
[17]:
ep.pl.tracksplot(ep_medcat, list(ep_medcat.anndata.var_names), groupby="Diabetes")